Introduction
Calixarenes are macrocycles, which feature a hydrophobic cavity. They are widely used for their ability to complex small molecules with potential biological applications. An extensive variety of structurally related molecules derived from calixarenes has been synthesized. This includes calix[
n]arenes with functionalized ligands.
[1] Since topological fragments required for building calix[
n]arenes with functionalized ligands are not available in the Cornell
et al. force field
[2] (and in its successive modifications as the additive Amber99SB force field) we developed a new Force Field Topology DataBase (FFTopDB) compatible with a selection of organic functions. The calix[
n]arenes studied in this work include oligomers of 2,4-dimethylaniline, 2,4-dimethylphenol, 2,4-dimethylthiophenol (with the corresponding anions), substituted or not at the C4 position by different alkyl groups (methyl, propyl, isopropyl, butyl, tertiobutyl, 1,1,2,2-tetramethylpropyl and allyl) and
via the heteroatom at the C1 position by various functionalized aliphatic ligands (with alcohol, ketone, ether, amine, amide, thiol, thioether, ester, and carboxylic acid organic functions). A calix[
n]arene aromatic unit is connected
via the C7 carbon to the C6 aromatic carbon belonging to the next (
n + 1) aromatic unit. 110 molecules were involved in charge derivation and the list of molecules consists of 12 molecules composing the calix[
n]arene aromatic rings, three
NH-methyl-,
O-methyl- and
S-methyl-based families of 26 ligands connected to the heteroatom at the C1 position and 8 alkyl derivatives connected at the C4 position (see Scheme a).
Computational details
Theory levels involved in quantum chemistry calculations were selected to be in agreement with these used in RESP charge derivation for the Amber99SB force field. The different building blocks used here were optimized using the HF/6-31G** theory level and the Firefly program (version 7.1.G).
[3] The lowest energy minimum for each aromatic building block was included in charge derivation. For the different alkyl molecules and functionalized ligands, one or two energy minima were selected after conformational search. A lowest minimum was considered only if no canonical intra-molecular hydrogen bond [donor (D)-acceptor (A) distance lower than 3.20 Å and the D-H...A angle between 120-180°] was observed in a structure. Molecular electrostatic potential (MEP) computation involved the Connolly surface algorithm and the HF/6-31G* theory level implemented in the Firefly program. For each aromatic moiety four molecular orientations and for each alkyl molecule and ligand, two molecular orientations based on the rigid-body reorientation algorithm implemented in R.E.D. program were involved in MEP computation assuring the reproducibility of charge values.
[4]
The molecular fragments required for MD simulations were constructed by setting intra-molecular charge constraints within the aromatic moieties and inter-molecular charge constraints between the aromatic moieties and the alkyl groups or ligands during the charge fitting step (see Scheme a). RESP charge fitting was carried out using a standalone version of the RESP program and following a two-stage fitting procedure with a hyperbolic restraint function, using a weighting factor of 0.0005 and 0.001 for the two stages, respectively.
[5] A Relative Root Mean Square (RRMS) value of 0.067 between the MEP calculated by quantum chemistry and that generated using the derived charge values was obtained for the last charge fitting step. A RRMS values of 0.068 was also obtained in the absence of intra- and inter-molecular charge constraints. The relative small RRMS values as well as the small difference of RRMS between the charge fitting steps carried out without and with intra- and inter-molecular charge constraints demonstrate the accuracy of the fitting step performed on 310 structures and the weak effect of the constraints used.
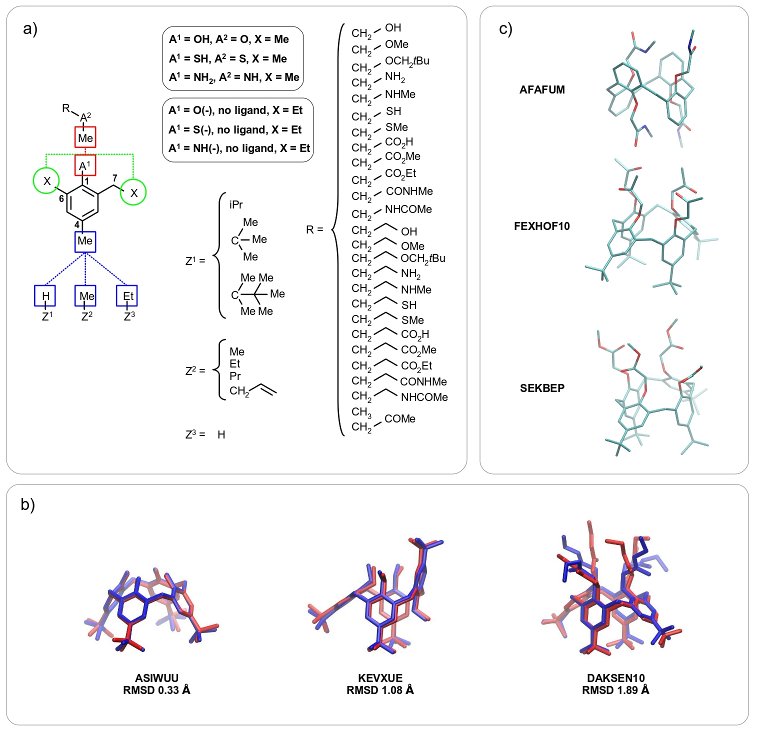 Scheme 1
Scheme 1
The new FFTopDB has been validated based on 10 nsec molecular dynamics (MD) simulations using structures originating from the
Cambridge Structural Database (CSD). The selected structures are referenced as ASIWUU
[6a], KEVXUE
[6b] and DAKSEN10
[6c] and are presented in scheme b. Averaged heavy atom Root Mean square Deviation (RMSD) values obtained between MD snapshots and CSD structures were computed for each model and was found to be 0.33, 1.08 and 1.89 Å for ASIWUU, KEVXUE and DAKSEN10, respectively. The flexibility of the ligands explains the relative high RMSD values obtained for KEVXUE and DAKSEN10. The RMSD values confirm the validity of the FFTopDB reported here.
Conclusion and new perpectives
Schemes b, c provide examples of calixarenes, which can be studied by MD simulations [calix[
4]arenes and calix[
n]arenes (with cone, partial cone and alternate conformations)]. A
LEaP script is available providing examples for building initial MD structures for substituted calix[
n]arenes. Finally, by directly defining new intra-molecular charge constraints set to the averaged 0.1494, 0.1768 and 0.1163 values obtained in this project for the methyl groups of the
NH-,
O- and
S-families of ligands, respectively, a potentially infinite number of new fragments (R-NH, R-O and R-S) can be added to the present list and could constitute add-ons to this present R.E.DD.B. project.
[1] Gutsche, C. David, Calixarenes, Cambridge, Royal Society of Chemistry, 1989.
[2] Cornell et al. J. Am. Chem. Soc. 1995, 117, 5179–5197.
[3] Nemukhin et al. Moscow Univ. Chem. Bull. 2004, 45, 75–102, and here.
[4] Information about the atoms involved in the rigid-body re-orientation algorithm is available in the PDB files of this R.E.DD.B. project (see the "REMARK REORIENT" keyword).
[5] Bayly et al. J. Phys. Chem. 1993, 97, 10269–10280, and here.
[6a] Asfari et al. Org. Biomol. Chem. 2005, 2, 387–396. [6b] Grootenhuis et al. J. Am. Chem. Soc. 1990, 112, 4165–4176. [6c] Arnaud-Neu et al. J. Am. Chem. Soc. 1989, 111, 8681–8691.









