This work represents an extension of the
F-71 R.E.DD.B. project (Gouin
et al. J. Org. Chem. 2007, 72, 9032). Eleven molecules [2-methoxyethanol, 2-(1-methyl-1H-1,2,3-triazol-4-yl)ethanol, (1-methyl-1H-1,2,3-triazol-4-yl)methanol, α
-D-glucose, β
-D-glucose, methyl α
-D-glucoside, methyl β
-D-glucoside, methyl α
-D-mannoside, methyl β
-D-mannoside, methyl α
-D-galactoside and methyl β
-D-galactoside] were involved in charge derivation and all-atom force field library building (Scheme 1A). The force field topology database (FFTopDB) generated here has been designed to be used in association with GLYCAM force field parameters (Scheme 1B). The R.E.D. program (in its version IV) provides for the first time the capabilities of charge derivation and force field library building for an entire set of molecules with fully characterized molecular orientations and conformations. In the present case, the derivation of atomic charges embedded in force field libraries for the different fragments constituting oligomeric organo-glycoconjugates was carried out in a single R.E.D. job. The corresponding FFTopDB has been generated in a single and fully consistent approach and is specific to the glycoclusters studied.
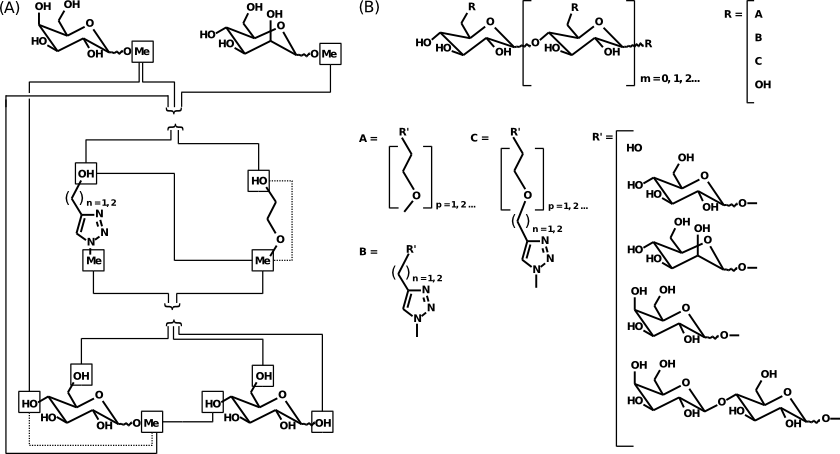 Scheme 1
Scheme 1
For each monosaccharide unit, two conformations based on the populations for the ω dihedral observed experimentally were considered: the
gt and
gg populations for the glucose and mannose derivatives, and the
gt and
tg populations for the galactose derivatives were selected. The lowest minimum found for (1-methyl-1H-1,2,3-triazol-4-yl)methanol and the two lowest minima observed for 2-methoxyethanol and 2-(1-methyl-1H-1,2,3-triazol-4-yl)ethanol after conformational search were chosen. Conformations presenting hydrogen bond(s) were excluded from the charge derivation procedure because they lead to over-polarization effect. Such an artifact is not suitable when using the additive force field model. Consequently, geometries of monosaccharides were optimized using dihedral angle constraints to avoid canonical hydrogen bonds. Nature and value of the constraints were the following: (i) the H4-C4-O4-HO4 dihedral of the
gt and
gg rotamers of α
-D-glucose and methyl α
-D-glucoside was constrained to the value of 180°, (ii) dihedrals H2-C2-O2-HO2 and H4-C4-O4-HO4 of the
gt and
gg rotamers of β
-D-glucose and methyl β
-D-glucoside were constrained to 180°, (iii) dihedrals H3-C3-O3-HO3 and H4-C4-O4-HO4 of the
gt and
gg rotamers of methyl α
-D-mannoside were constrained to 60 and 180°, respectively, (iv) dihedral H2-C2-O2-HO2 of the
gt rotamer of methyl β
-D-mannoside was constrained to -30°, while dihedrals H2-C2-O2-HO2 and H3-C3-O3-HO3 of the
gg rotamer were constrained to -30 and 30°, respectively, (v) dihedrals H2-C2-O2-HO2 and H3-C3-O3-HO3 of the
gt rotamer of methyl α
-D-galactoside were constrained to -60°, while dihedrals H2-C2-O2-HO2, H3-C3-O3-HO3 and H4-C4-O4-HO4 of the
tg rotamer were constrained to -60, 60 and 30°, respectively, (vi) dihedrals H2-C2-O2-HO2, H3-C3-O3-HO3 and H4-C4-O4-HO4 of the
gt rotamer of methyl β
-D-galactoside were constrained to 180, -60 and -60°, respectively, while dihedrals H3-C3-O3-HO3 and H4-C4-O4-HO4 of the
tg rotamer were constrained to -30° and 30°, respectively.
Geometry optimization and molecular electrostatic potential (MEP) computation were carried out using the Gaussian 03 program, while charge fitting was performed using the RESP program. The Hartree-Fock method and the 6-31G** basis set were used in geometry optimization. MEP computation was carried out using the HF/6-31G* theory level and the CHELPG algorithm after control of the molecular orientation of each optimized geometry using the rigid-body reorientation algorithm implemented in the R.E.D. IV program. For each optimized geometry, four orientations [based on the C1, C3, C5; C5, C3, C1; C2, C4, O5; O5, C4, C2 sets of three atoms for each monosaccharide unit; the CX, O1, C2; C2, O1, CX; C2, C3, OY; OY, C3, C2 atoms for 2-methoxyethanol; the OY, C1, C2; C2, C1, OY; C2, C3, N4; N4, C3, C2 atoms for 2-(1-methyl-1H-1,2,3-triazol-4-yl)ethanol, and the OY, C1, C2; C2, C1, OY; C2, N5, C6; C6, N5, C2 atoms for (1-methyl-1H-1,2,3-triazol-4-yl)methanol] were computed (atom names are described in the PDB files of this project).
A single RESP stage and a hyperbolic constraint value of 1.0E-2 was used during the charge fitting step. Molecular fragments were designed by combining various intra- and inter-molecular charge constraints set to a value of zero during the fit. Intra-molecular charge constraints were defined (i) between the hydroxyl and methyl groups of 2-methoxyethanol, (ii) between the HO4 hydroxyl and the methyl groups of methyl α and β
-D-glucosides, and (iii) for each hydrogen bound to a sp3 carbon. Inter-molecular charge constraints were defined between (i) the hydroxyl group of each triazole unit and the methyl group of 2-methoxyethanol, (ii) the hydroxyl group of 2-methoxyethanol and the methyl group of each methyl glycoside, (iii) the hydroxyl group of each triazole unit and the methyl group of each methyl glycoside, (iv) the HO4 hydroxyl group of methyl α- or β
-D-glucoside and the methyl group of methyl β
-D-galactoside, (v) the HO4 hydroxyl group of α- or β
-D-glucose and the methyl group of methyl α- or β
-D-glucoside, (vi) the HO1 or HO6 hydroxyl group of α- or β
-D-glucose and the methyl group of 2-methoxyethanol, and (vii) the HO1 or HO6 hydroxyl group of α- and β
-D-glucose and the methyl group of each triazole unit. Inter-molecular charge equivalencing for the O1 atom of 2-methoxyethanol and methyl glycosides was also applied.
A RRMS (relative root mean square value between the MEP computed by quantum mechanics, and that calculated using the derived charge values) of 0.126 was obtained for the fit. Intra-, inter-molecular charge constraints and inter-molecular charge equivalencing are responsible of an increase of 0.009 of the RRMS value. The low RRMS values and the relative small difference between the RRMS calculated without and with the charge constraints demonstrate the effectiveness of the approach. The glycocluster FFTopDB is available as a suite of force field libraries for molecular fragments in the Tripos mol2 file format and is used to build Amber OFF force field libraries. A LEaP script is available for this purpose in this R.E.DD.B. project. The corresponding charge set has been validated and the corresponding study will be included in a forthcoming paper.









