Introduction
We report on a new force field topology database (FFTopDB;
i.e. RESP charges embedded in a set of 89 force field libraries in the
Tripos mol2 and
mol3 file formats) for more than 200 biochemical cofactors and vitamins involved in numerous biological processes. This FFTopDB is compatible with the Duan
et al. force field,
[1] and is devoted to condensed phase molecular dynamics simulations and docking studies. The non-exhaustive list of cofactors presented here contains non-phosphorylated and phosphorylated (phosphate and hydrogen-phosphate) derivatives such as:
- X (natural nucleosides and 2'-Deoxynucleosides),
- XYP (Adenosine monophosphate, Adenosine diphosphate, Adenosine triphosphate, ...),
- cyclic-XMP (cyclic AMP, ...),
- NXD+ (Nicotinamide adenine dinucleotide, ...),
- NXDH (the reduced form of NAD+, ...),
- NXDYP (Nicotinamide adenine dinucleotide phosphate, ...),
- NXDYPH (the reduced form of NADP, ...),
- riboflavin,
- FMN (Flavin mononucleotide),
- FMNH2 (the reduced form of Flavin mononucleotide),
- FXD (Flavin adenine dinucleotide, ...),
- FXDH2 (the reduced form of FAD, ...),
- FXDYP (Flavin adenine dinucleotide phosphate, ...),
- FXDYPH2 (the reduced form of FADP, ...),
- acetylated and non-acylated coenzyme XYP (Acetyl Coenzyme A, ...);
with X = 2'-Deoxyadenosine, 2'-Deoxycytidine, 2'-Deoxyguanosine, 2'-Deoxythymidine, Adenosine, Cytidine, Guanosine or Uridine, and Y = any positive integer value.
[2]
In this work, the "building block" procedure has been followed: charge derivation and force field library building were performed by using well-defined elementary constituents for the different studied cofactors. Thus, the entire family of cofactors is considered as a single homogeneous biopolymer model in our approach (Figure 1A).
[3] Specific charge constraints for fully characterized connecting groups belonging to these building blocks were applied during the charge fitting step allowing the generation of a large number of molecular fragments. The cofactors are then constructed by associating the molecular fragments together by using a dedicated LEaP
script. The building block approach presents the following advantages over the whole molecule approach: (i) the cpu time required for geometry optimization and molecular electrostatic potential (MEP) computation is drastically decreased, (ii) the optimized geometry of the conformation(s) of each building block is fully defined and controlled, (iii) conformations not suited for charge derivation, presenting non-bonded interactions only observed in gas phase geometry optimization are discarded, (iv) cofactors and their analogs are simultaneously involved in a single and highly homogenous approach, and finally (v) by generating averaged charge values for connecting groups, additional and highly compatible charge derivation procedures can be performed for an infinity of new cofactor analogs constituting "add-ons" to the present R.E.DD.B. project.
Computational details
The charge derivation procedure and force field library building were automatically carried out by using the R.E.D. IV program allowing a rigorous control of the different parameters, which affect charge values compatible with the non-polarizable RESP charge model.
[4] Geometry optimization and MEP computation were carried out by quantum mechanical methods by using the
Gaussian 2003 (version E.01) program. The geometries of the 28 building blocks considered in this work were optimized using the HF/6-31G* level of theory.
[5] One to four molecular conformations were used for each building block depending on their occurrence in the
protein data bank.
[6] An energy minimum was considered only if no canonical intra-molecular hydrogen bond [donor (D)-acceptor (A) distance lower than 3.20 Å and the D-H...A angle between 120-180°] was observed in each optimized geometry. Dihedral constraints were used in geometry optimization to prevent intra-molecular hydrogen bond formation when needed.
[7] MEP computation employed the B3LYP/cc-pVTZ level of theory, the Polarized Continuum Model - the Integral Equation Formalism mimicking the diethylether environment, and the Connolly surface algorithm defined in the Duan et al. force field.
[1] For each building block one or two pairs of molecular orientations based on the rigid-body reorientation algorithm (RBRA) implemented in the R.E.D. program were considered in MEP computation ensuring the reproducibility of the derived charge values.
[8] A total of 89 molecular fragments were generated by setting specific intra- and inter-molecular charge constraints between the connecting groups during the charge fitting step (see Figure 1A). Inter-molecular charge equivalencing was used to force the atomic charges between the common elements of the different building blocks to be equivalent leading to a highly consistent set of charge values within the FFTopDB. In this approach, the charges of the C1'/H1' and N1 or N9 connecting atoms between 2'-deoxyribose/ribose and each nucleobase as well as these of the ribose carbons and hydrogens of the nicotinamide nucleoside (oxidized form) were excluded from these constraints to limit the impact on the Relative Root Mean Square (RRMS).
[9] RESP charge fitting was carried out by using a standalone version of the RESP program, and following the two RESP stage fitting procedure.
[10]
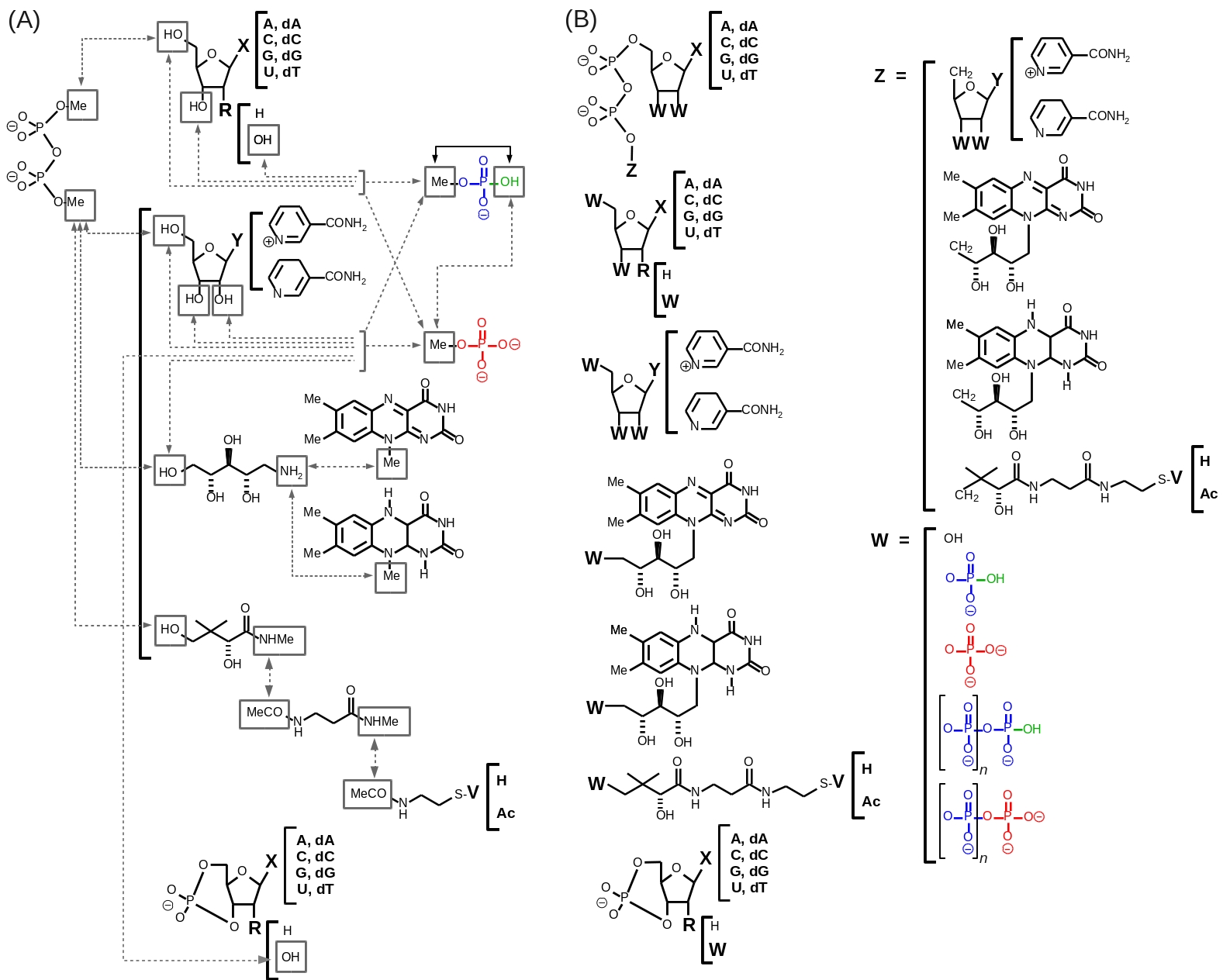 Figure 1
Figure 1
The statistics module of the R.E.D. IV program was used to minimize the impact of the intra-molecular charge constraints, inter-molecular charge constraints and inter-molecular charge equivalencing in the charge fitting step for multiple molecules. A RRMS value of 0.026 between the MEP calculated by quantum chemistry and that generated using the derived charge values was obtained for the charge fitting step. A highly similar RRMS value was also obtained in the absence of intra-molecular charge constraints, inter-molecular charge constraints and inter-molecular charge equivalencing. The relative small RRMS values as well as the small difference of RRMS between the charge fitting steps carried out with and without these charge constraints is one of the ways to demonstrate the accuracy of the fitting step performed in this work and the relative weak effect of the constraints used. Finally, rounding off errors of charge values were corrected at the fourth decimal point. RESP charges were validated by molecular dynamics simulations in condensed phase conditions.
This R.E.DD.B. project provides all the computational conditions for charge derivation and force field library building for more than 200 biochemical cofactors. Moreover, this allows any user to rebuild the FFTopDB by applying other choices (different conformations and orientations, different algorithms in MEP computation or ESP charge fitting). A similar FFTopDB compatible with the Cornell
et al. force field is also available in the "
F-90" R.E.DD.B. project.
[11] FFTopDBs for these cofactors with extra-points and/or united carbon atoms are also in preparation.
[1] Duan
et al. J. Comput. Chem. 2003,
24, 1999–2012.
[2] Wikipedia,
biochemical cofactors.
[3] Cieplak
et al. J. Comput. Chem. 1995,
16 1357–1377.
[4] Dupradeau
et al. Phys. Chem. Chem. Phys. 2010,
12, 7821–7839.
[5] The names for the 28 building blocks considered in this work are in the order: Dimethyldiphosphate; Methyl-hydrogen-phosphate; Methyl-phosphate; 2'-Deoxyadenosine; 2'-Deoxycytidine; 2'-Deoxyguanosine; 2'-Deoxythymidine; Adenosine; Cytidine; Guanosine; Uridine; 1-D-ribofuranosyl-1,4-dihydropyridine-3-carboxamide; 3-(aminocarbonyl)-1-D-ribofuranosylpyridinium; (2S,3S,4R)-1-amino-1-deoxypentitol; 7,8,10-trimethylbenzo[g]pteridine-2,4(3H,10H)-dione; 7,8,10-trimethyl-5,10-dihydrobenzo[g]pteridine-2,4(1H,3H)-dione; (2R)-2,4-dihydroxy-N,3,3-trimethylbutanamide; N3-acetyl-N-methyl-b-alaninamide; S-[2-(acetylamino)ethyl]-ethanethioate; N-(2-sulfanylethyl)acetamide; 2'-Deoxy-3',5'-cyclic-adenosine-monophopshate; 2'-Deoxy-3',5'-cyclic-cytidine-monophosphate; 2'-Deoxy-3',5'-cyclic-guanosine-monophosphate; 2'-Deoxy-3',5'-cyclic-thymidine-monophosphate; 3',5'-cyclic-adenosine-monophosphate; 3',5'-cyclic-cytidine-monophosphate; 3',5'-cyclic-guanosine-monophosphate; 3',5'-cyclic-uridine-monophosphate.
[6] Berman
et al. Nucl. Acids Res. 2000,
28, 235–242.
[7] The dihedrals, which were constrained during the geometry optimization step are described as it follows: molecule name: total number of constrained dihedral(s); four atom numbers defining the dihedral, the value of the dihedral constraint: Adenosine (conformations C3'endo & C2'endo): 2; 4 3 9 11, 180.0; 9 7 17 18, -70.0; Cytidine (conformations C3'endo & C2'endo): 2; 4 3 9 11, 180.0; 9 7 17 18, -70.0; Guanosine (conformations C3'endo & C2'endo): 2; 4 3 9 11, 180.0; 9 7 17 18, -70.0; Uridine (conformations C3'endo & C2'endo): 2; 4 3 9 11, 180.0; 9 7 17 18, -70.0; 1-D-ribofuranosyl-1,4-dihydropyridine-3-carboxamide (conformations C3'endo/anti, C2'endo/anti, C2'endo/syn): 2; 4 3 9 11, 180.0; 9 7 17 18, -70.0; 1-D-ribofuranosyl-1,4-dihydropyridine-3-carboxamide (conformation C3'endo/syn): 3; 2 1 13 11, 180.0; 4 3 9 11, 180.0; 9 7 17 18, -70.0; 3-(aminocarbonyl)-1-D-ribofuranosylpyridinium (conformations C3'endo/anti, C3'endo/syn, C2'endo/anti, C2'endo/syn): 2; 4 3 9 11, 180.0; 9 7 17 18, -70.0; (2S,3S,4R)-1-amino-1-deoxypentitol; 2; 8 7 9 10, 30.0; 16 15 17 18, -170.0; 3',5'-cyclic-adenosine-monophosphate; 1; 10 8 18 19, -70.0; 3',5'-cyclic-cytidine-monophosphate; 1; 10 8 18 19, -70.0; 3',5'-cyclic-guanosine-monophosphate; 1; 10 8 18 19, -70.0; 3',5'-cyclic-uridine-monophosphate; 1; 10 8 18 19, -70.0.
[8] Atoms involved in the RBRA procedure before MEP computation are defined as it follows: molecule number (1-28): total number of molecular orientations; three atom numbers separated by the pipe character: 1: 4; 5 9 13 | 13 9 5 | 6 9 10 | 10 9 6; 2: 2; 1 5 6 | 6 5 1; 3: 2; 1 5 6 | 6 5 1; 4: 4; 5 10 14 | 14 10 5 | 7 12 17 | 17 12 7; 5: 4; 5 10 14 | 14 10 5 | 7 12 17 | 17 12 7; 6: 4; 5 10 14 | 14 10 5 | 7 12 17 | 17 12 7; 7: 4; 5 10 14 | 14 10 5 | 7 12 17 | 17 12 7; 8: 4; 5 9 13 | 13 9 5 | 7 11 16 | 16 11 7; 9: 4; 5 9 13 | 13 9 5 | 7 11 16 | 16 11 7; 10: 4; 5 9 13 | 13 9 5 | 7 11 16 | 16 11 7; 11: 4; 5 9 13 | 13 9 5 | 7 11 16 | 16 11 7; 12: 4; 5 9 13 | 13 9 5 | 7 11 16 | 16 11 7; 13: 4; 5 9 13 | 13 9 5 | 7 11 16 | 16 11 7; 14: 2; 4 7 11 | 11 7 4; 15: 4; 9 10 12 | 12 10 9 | 1 4 8 | 8 4 1; 16: 4; 10 11 14 | 14 11 10 | 1 5 9 | 9 5 1; 17: 2; 3 6 15 | 15 6 3; 18: 2; 9 12 15 | 15 12 9; 19: 2; 9 12 15 | 15 12 9; 20: 2; 9 12 15 | 15 12 9; 21: 4; 6 11 15 | 15 11 6 | 8 13 18 | 18 13 8; 22: 4; 6 11 15 | 15 11 6 | 8 13 18 | 18 13 8; 23: 4; 6 11 15 | 15 11 6 | 8 13 18 | 18 13 8; 24: 4; 6 11 15 | 15 11 6 | 8 13 18 | 18 13 8; 25: 4; 6 10 14 | 14 10 6 | 8 12 17 | 17 12 8; 26: 4; 6 10 14 | 14 10 6 | 8 12 17 | 17 12 8; 27: 4; 6 10 14 | 14 10 6 | 8 12 17 | 17 12 8; 28: 4; 6 10 14 | 14 10 6 | 8 12 17 | 17 12 8.
[9] Inter-molecular charge equivalencing between building blocks applied during the charge fitting stepare are defined as it follows: molecule numbers | atom numbers in the set of molecules: 4 5 6 7 | 1 2 3 4 7 8 9 10 11 12 13 14 15 16 17; 8 9 10 11 12 13 | 1 2 3 4 17 1; 8 9 10 11 12 | 7 8 9 10 11 12 13 14 15 16; 21 22 23 24 | 1 2 3 4 5 8 9 10 11 12 13 14 15 16 17 18; 25 26 27 28 | 1 2 3 4 5 8 9 10 11 12 13 14 15 16 17 18 19; 4 8 21 25 | 18 19 19 20 - 19 20 20 21 - 20 21 21 22 - 21 22 22 23 - 22 23 23 24 - 23 24 24 25 - 24 25 25 26 - 25 26 26 27 - 27 28 28 29 - 28 29 29 30 - 29 30 30 31 - 30 31 31 32 - 31 32 32 33; 5 9 22 26 | 18 19 19 20 - 19 20 20 21 - 20 21 21 22 - 21 22 22 23 - 22 23 23 24 - 23 24 24 25 - 24 25 25 26 - 26 27 27 28 - 27 28 28 29 - 28 29 29 30 - 29 30 30 31; 6 10 23 27 | 18 19 19 20 - 19 20 20 21 - 20 21 21 22 - 21 22 22 23 - 22 23 23 24 - 23 24 24 25 - 24 25 25 26 - 25 26 26 27 - 26 27 27 28 - 28 29 29 30 - 29 30 30 31 - 30 31 31 32 - 31 32 32 33 - 32 33 33 34; 7 24 | 18 19 - 19 20 - 20 21 - 21 22 - 22 23 - 23 24 - 25 26 - 26 27 - 27 28 - 28 29 - 29 30 - 30 31 - 31 32; 11 28 | 19 20 - 20 21 - 21 22 - 22 23 - 23 24 - 24 25 - 26 27 - 27 28 - 28 29 - 29 30.
[10] Bayly
et al. J. Phys. Chem. 1993,
97, 10269–10280, and
here.
[11] Cornell
et al. J. Am. Chem. Soc. 1995,
117, 5179–5197; Hornak
et al. Proteins 2006,
65, 712–725.









