

RESP ESP charge Derive
Home Page
R.E.D. allows computational biologists involved in AMBER, CHARMM, GLYCAM & OPLS force field based biological studies to automatically derive RESP and ESP charge values, and to build force field libraries for new molecules/molecular fragments.
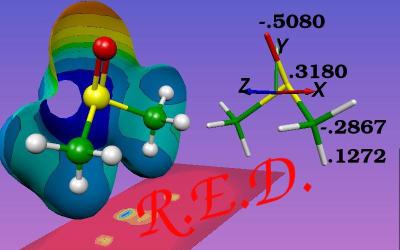 Done using Molekel...
Done using Molekel...
R.E.D. version III.5x
. R.E.D.-III tools release: April 2007
. R.E.D.-III.1 tools release: January 2009
. R.E.D.-III.2 tools release: April 2009
. R.E.D.-III.3 tools release: October 2009
. R.E.D.-III.4 tools release: July 2010
. R.E.D.-III.5 tools release: February 2012
UNIX, Mac OS X & Windows compatible
|
Done using Jmol &
Java (JRE) -
Help about installing JRE. Problems with Java? Use Pale Moon instead of Firefox or Chrome. |
|
|
Last update of the R.E.D. Home Page: June 16th, 2017. |